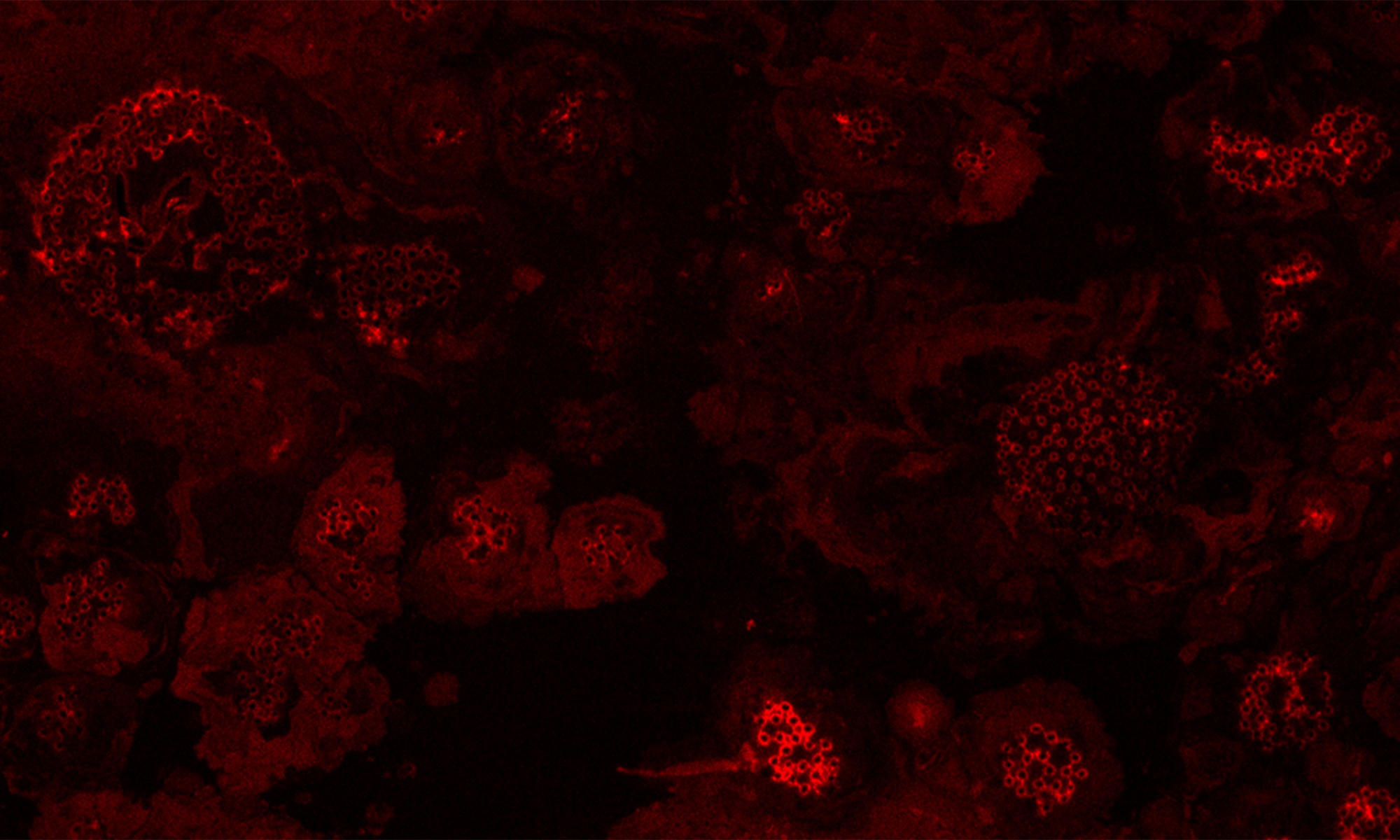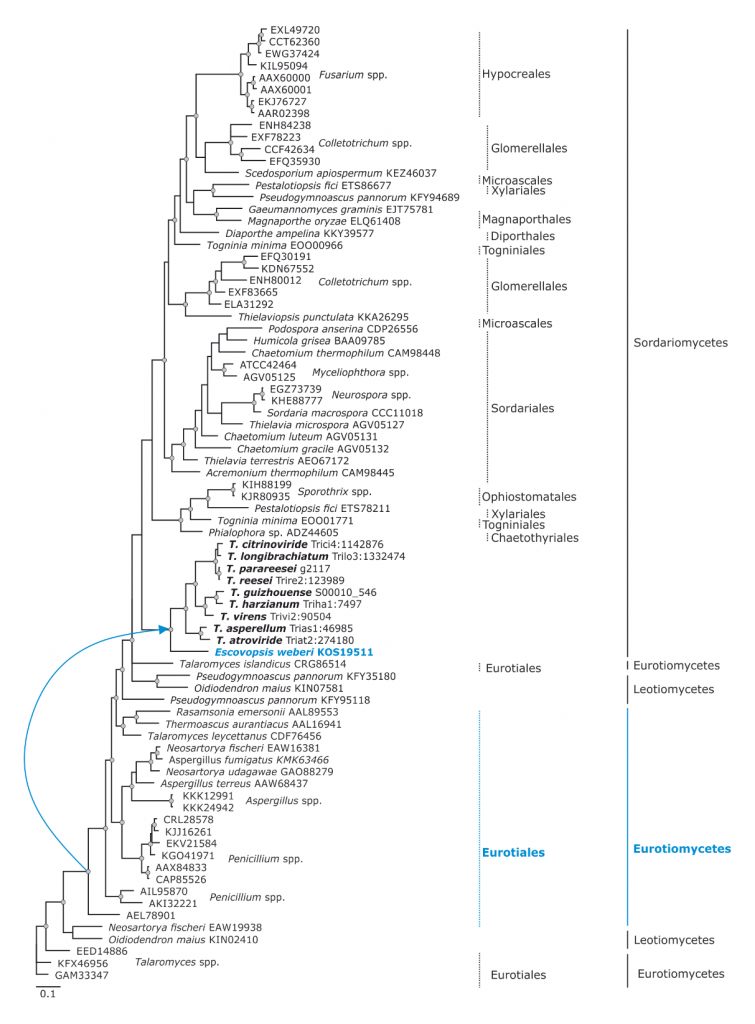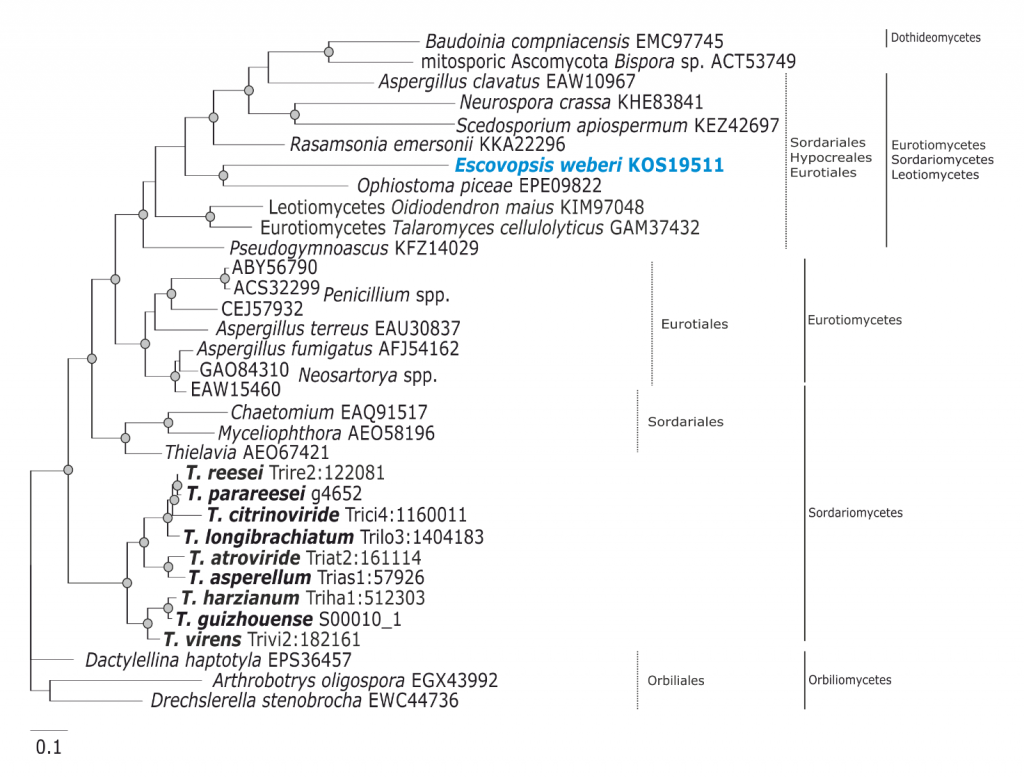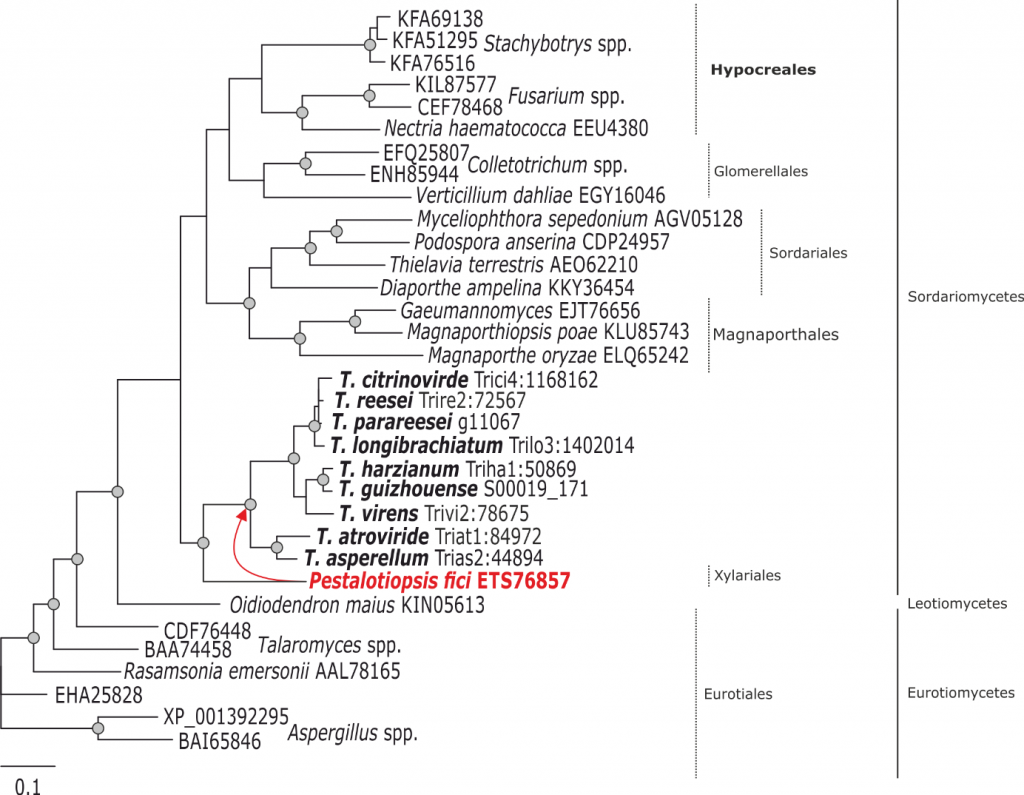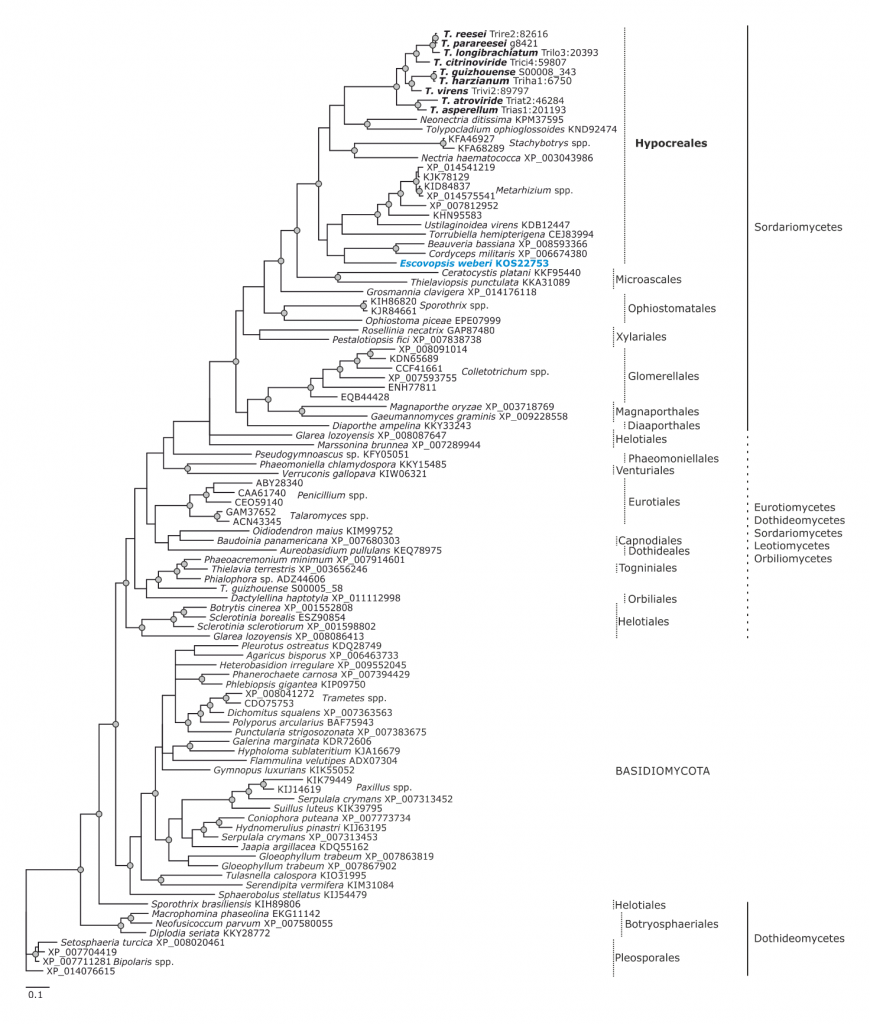
Phylogram based on Dayhoff amino acid substitution model using an alignment containing 70686 characters. Bayesian analysis was run for 1 million mcmc generations and a strict consensus tree was obtained by summarizing 7500 trees, after burning first 25% of obtained 10,000 trees. Mean tree length and variance are 2.425237E+01 and 7.0550690E-02, respectively. Posterior probabilities more than 95% are marked with circular nodes. Dashed vertical bars and non-dashed vertical bars represents the taxonomic order and taxonomic class in the phylum Ascomycota, respectively