Ding, M., Chen, W., Gao, R., Jiang, S., Zhao, Z., Cai, F., and Druzhinina, I. S. Emerging salt marshes as a source of Trichoderma arenerea sp. nov. and other fungal bioeffectors for biosaline agriculture, Journal of Applied Microbiology, doi: 10.1111/jam.14751.
Aims: Sustainable agriculture requires effective and safe biofertilizers and biofungicides with low environmental impact. Natural ecosystems that closely resemble the conditions of biosaline agriculture may present a reservoir for fungal strains that can be used as novel bioeffectors.
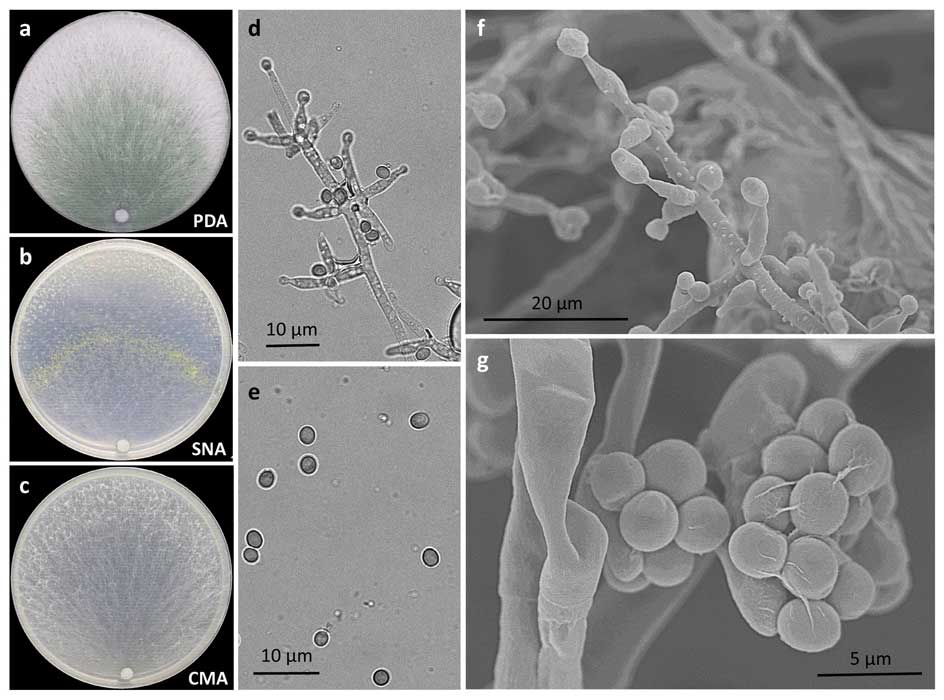
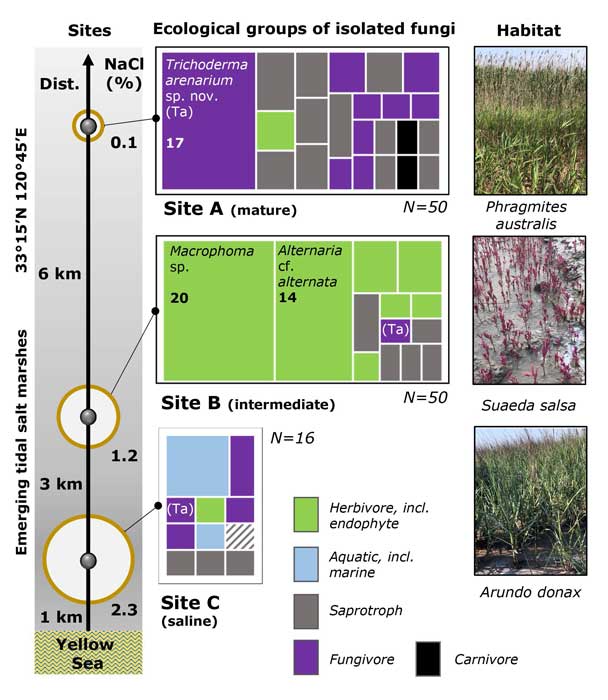
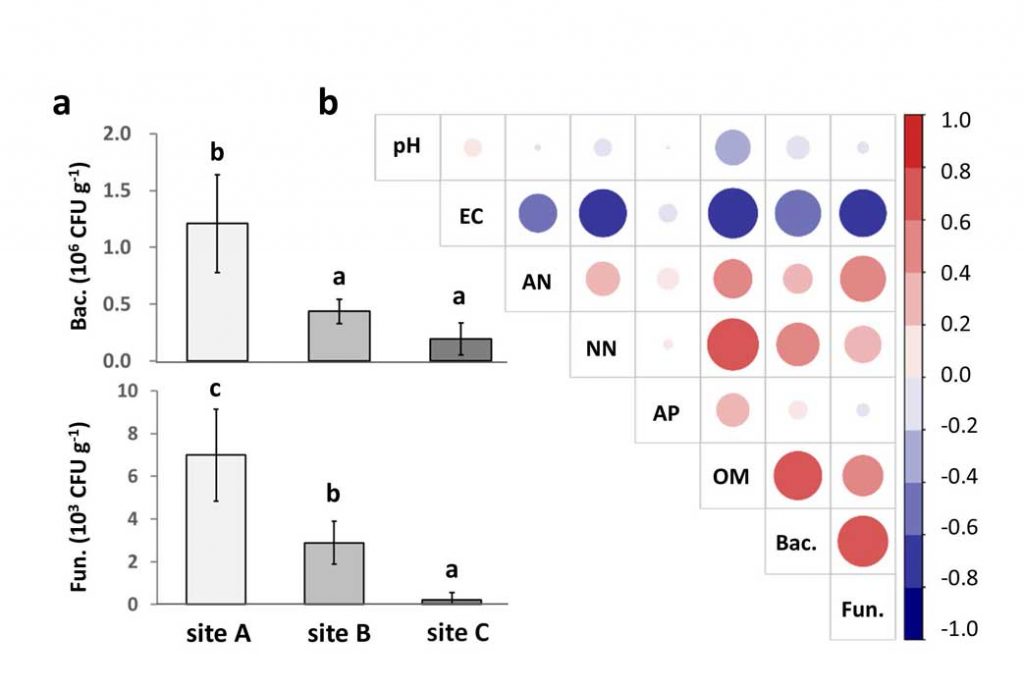
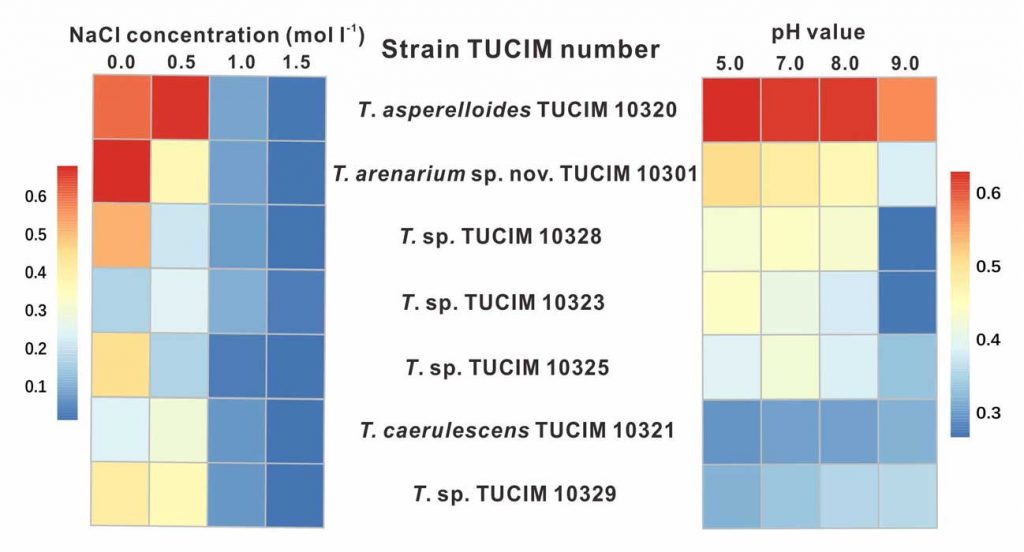
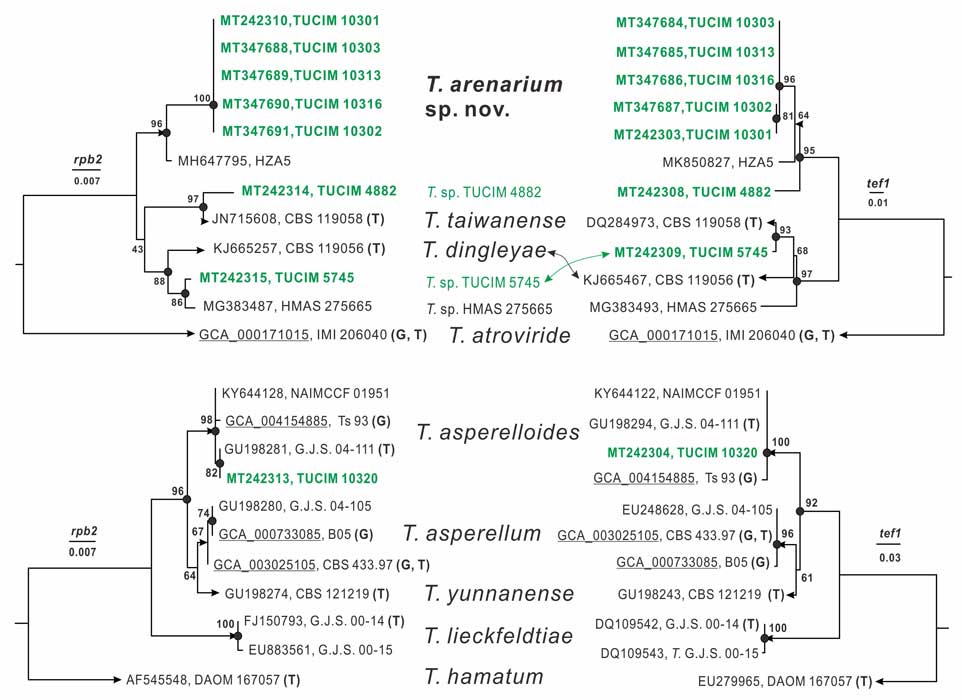
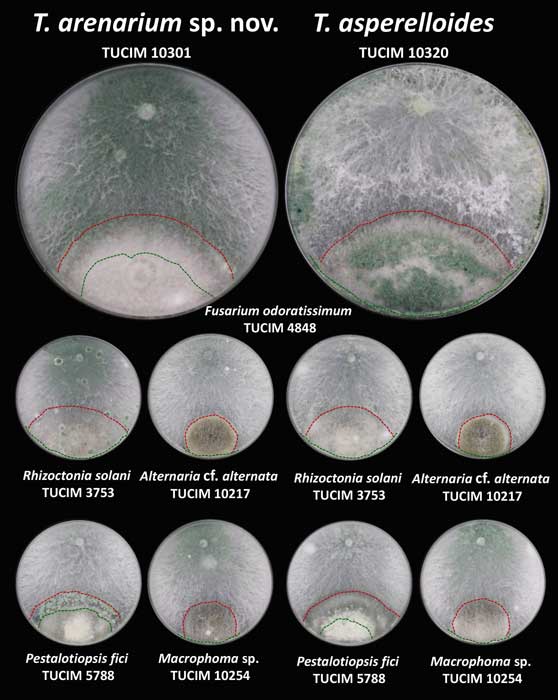
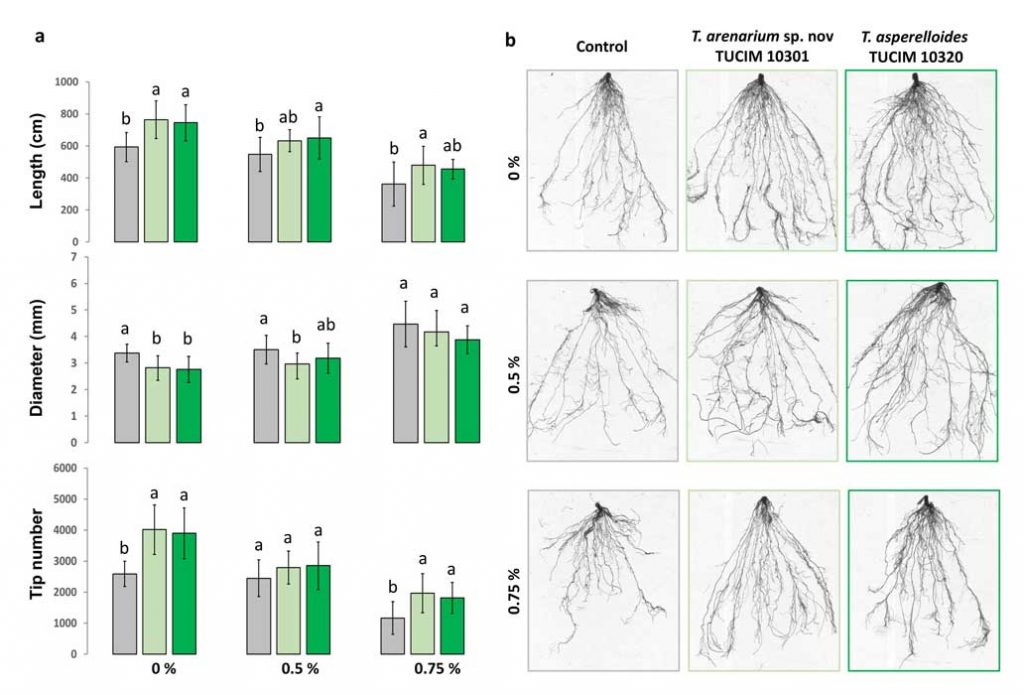
Methods and results: We isolated a library of fungi from the rhizosphere of three natural halotolerant plants grown in the emerging tidal salt marshes on the southeast coast of China. DNA barcoding of 116 isolates based on the rRNA ITS1 and 2 and other markers (tef1 or rpb2) revealed 38 fungal species, including plant pathogenic (41%), saprotrophic (24%), and mycoparasitic (28%) taxa. The mycoparasitic fungi were mainly species from the hypocrealean genus Trichoderma, including at least four novel phylotypes. Two of them, representing the taxa Trichoderma arenarium sp. nov. (described here) and T. asperelloides, showed effective antagonistic activity against five phytopathogenic fungi, and significant growth promotion on tomato seedlings under the conditions of saline agriculture.
Conclusions: Trichoderma spp. of salt marshes play the role of natural biological control in young soil ecosystems with a putatively premature microbiome.
Significance and impact: The saline soil microbiome is a rich source of halotolerant bioeffectors that can be used in biosaline agriculture.
Keywords: Trichoderma arenarium; Biosaline agriculture; Halotolerant fungi; Plant growth promotion; Rhizosphere; Salt marsh.