
Welcoming the Year of the Ox


Virtual fungal genomics laboratory of Irina S. Druzhinina and Feng Cai








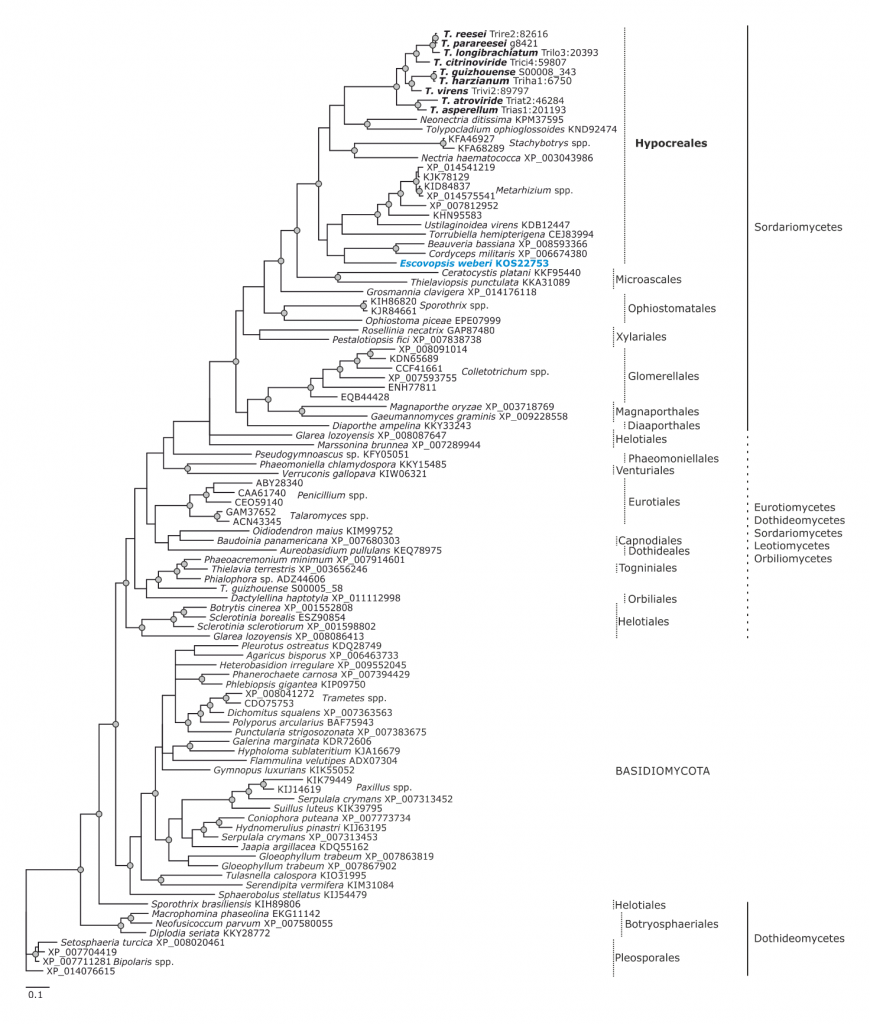
Phylogram based on Dayhoff amino acid substitution model using an alignment containing 70686 characters. Bayesian analysis was run for 1 million mcmc generations and a strict consensus tree was obtained by summarizing 7500 trees, after burning first 25% of obtained 10,000 trees. Mean tree length and variance are 2.425237E+01 and 7.0550690E-02, respectively. Posterior probabilities more than 95% are marked with circular nodes. Dashed vertical bars and non-dashed vertical bars represents the taxonomic order and taxonomic class in the phylum Ascomycota, respectively
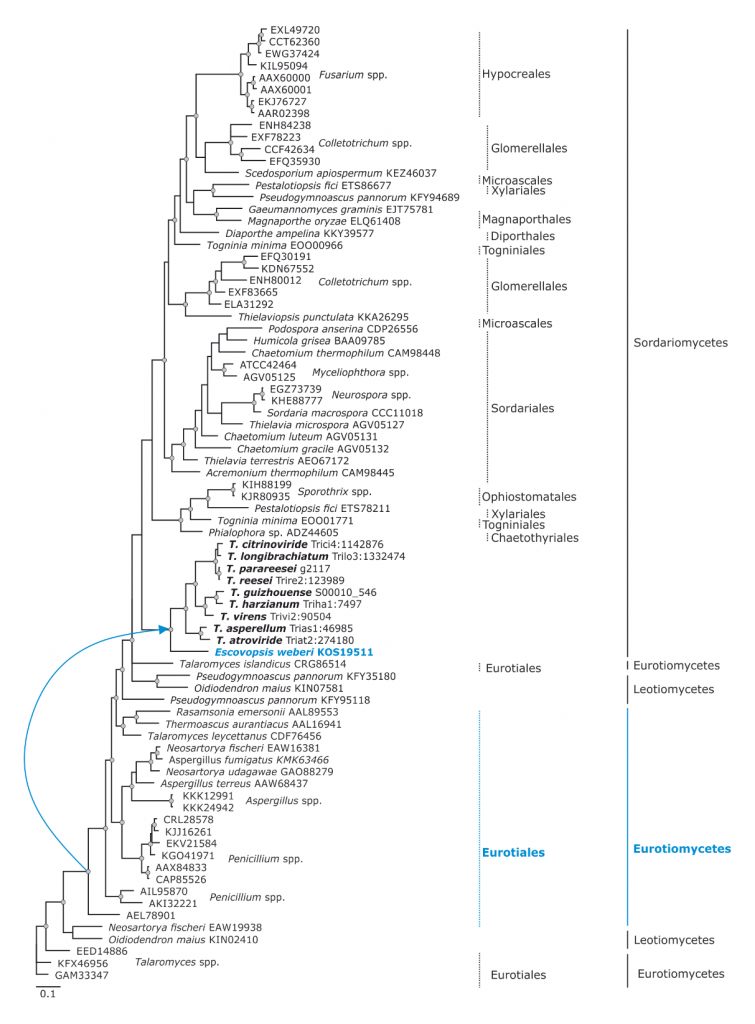
Phylogram based on Dayhoff amino acid substitution model using an alignment containing 38720 characters. Bayesian analysis was run for 1 million mcmc generations and a strict consensus tree was obtained by summarizing 7500 trees, after burning first 25% of obtained 10,000 trees. Mean tree length and variance are 9.480271E+00 and 2.2693320E-02, respectively. Posterior probabilities more than 95% are marked with circular nodes. Dashed vertical bars and non-dashed vertical bars represents the taxonomic order and taxonomic class in the phylum Ascomycota, respectively. LGT inferred by phylogeny and NOTUNG is shown with a blue arrow since it is occuring before Trichoderma and Escovopsis diverged and the respective donors are marked in blue.
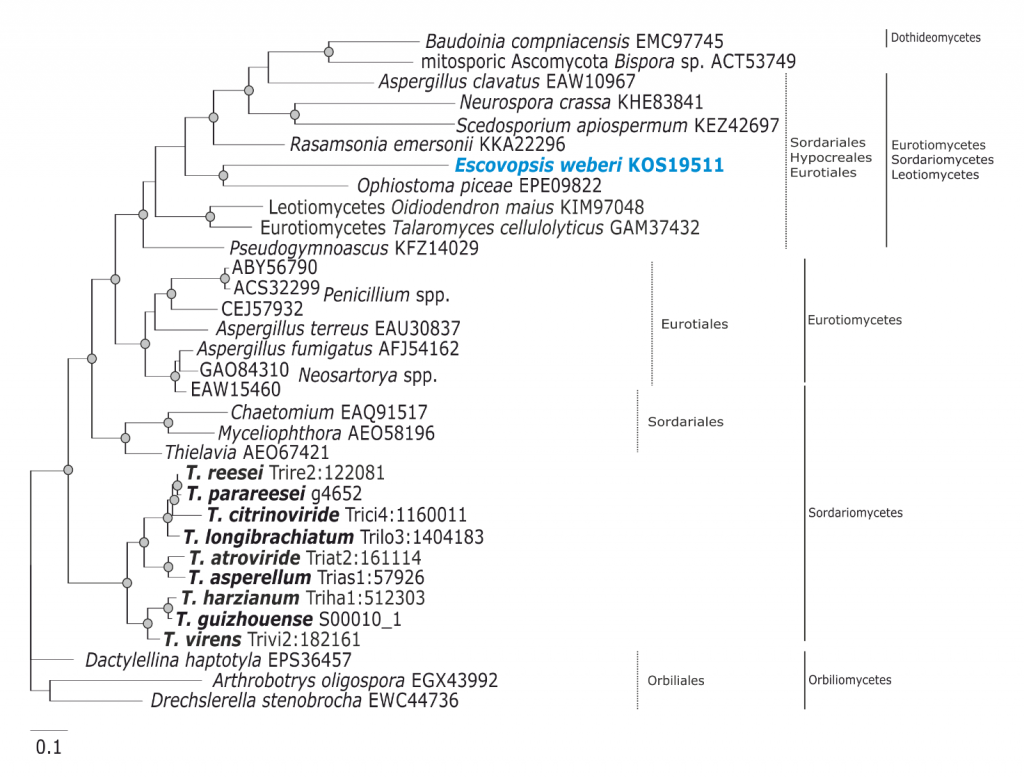
Phylogram based on Dayhoff amino acid substitution model using an alignment containing 18546 characters. Bayesian analysis was run for 1 million mcmc generations and a strict consensus tree was obtained by summarizing 7500 trees, after burning first 25% of obtained 10,000 trees. Mean tree length and variance are 7.457580E+00 and 2.1536720E-02, respectively. Posterior probabilities more than 95% are marked with circular nodes. Dashed vertical bars and non-dashed vertical bars represents the taxonomic order and taxonomic class in the phylum Ascomycota, respectively
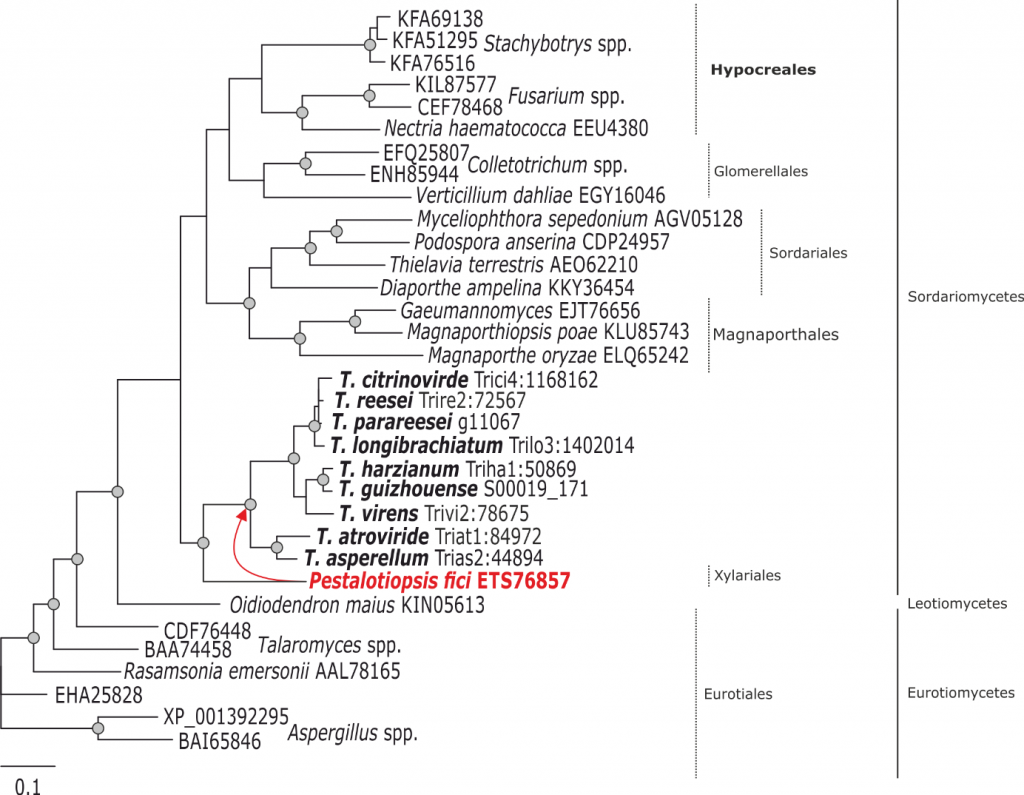
Phylogram based on Dayhoff amino acid substitution model using an alignment containing 20707 characters. Bayesian analysis was run for 1 million mcmc generations and a strict consensus tree was obtained by summarizing 7500 trees, after burning first 25% of obtained 10,000 trees. Mean tree length and variance are 7.269450E+00 and 3.9080650E-02, respectively. Posterior probabilities more than 95% are marked with circular nodes. Dashed vertical bars and non-dashed vertical bars represents the taxonomic order and taxonomic class in the phylum Ascomycota, respectively. LGT inferred by NOTUNG and Phylogeny is shown with a red arrow to the node and the respective donor is marked in red.
December 25, 2020: The Christmas talk “Fungal surface-active proteins unveil secrets of the tubular lifestyle, fitness, and speciation” at the academic meeting of the State Key Laboratory of Microbial Metabolism in School of Life Sciences and Biotechnology, Shanghai Jiao Tong University The meeting was organized by Prof. Jie Chen (see image above)





Prof. Chengshu Wang’s Lab in the Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Science, Shanghai is studying entomopathogenic fungi and fungal-insect interactions. Prof. Sibao Wang from te same Center gave us an introduction to the mosquito microbiome research.




An interactive course on fungal biology:
Today we had a graphic test on ultrastructure of fungal cells. Each student had 1 m of hypha to draw and annotate. Fortunately, we have a long bench in the corridor.




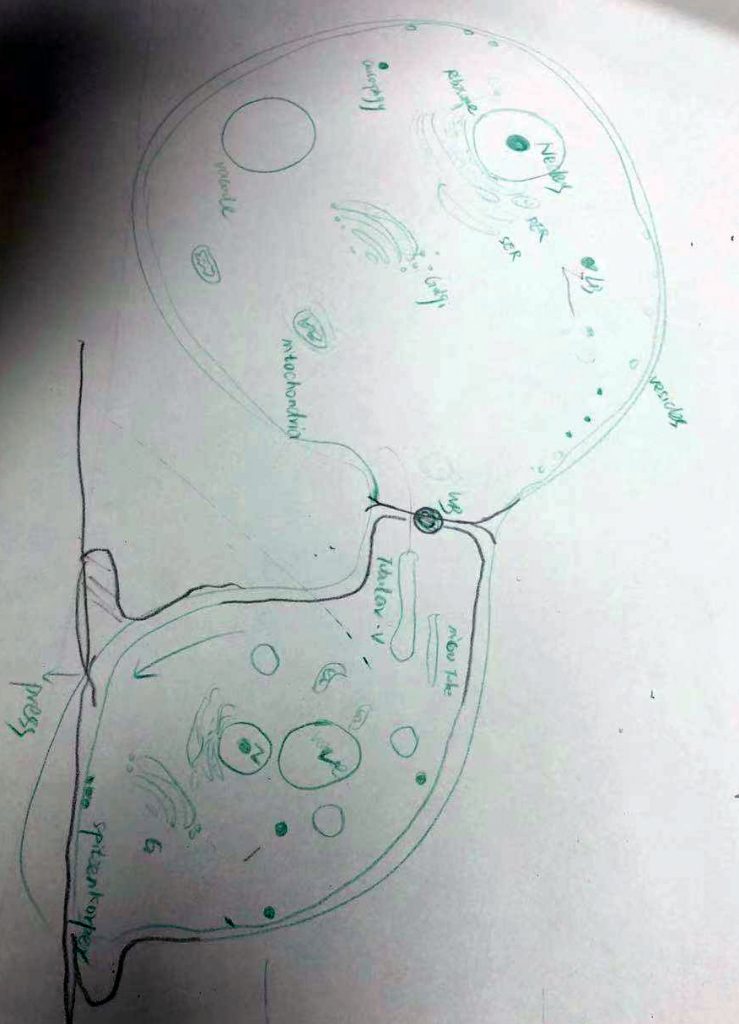

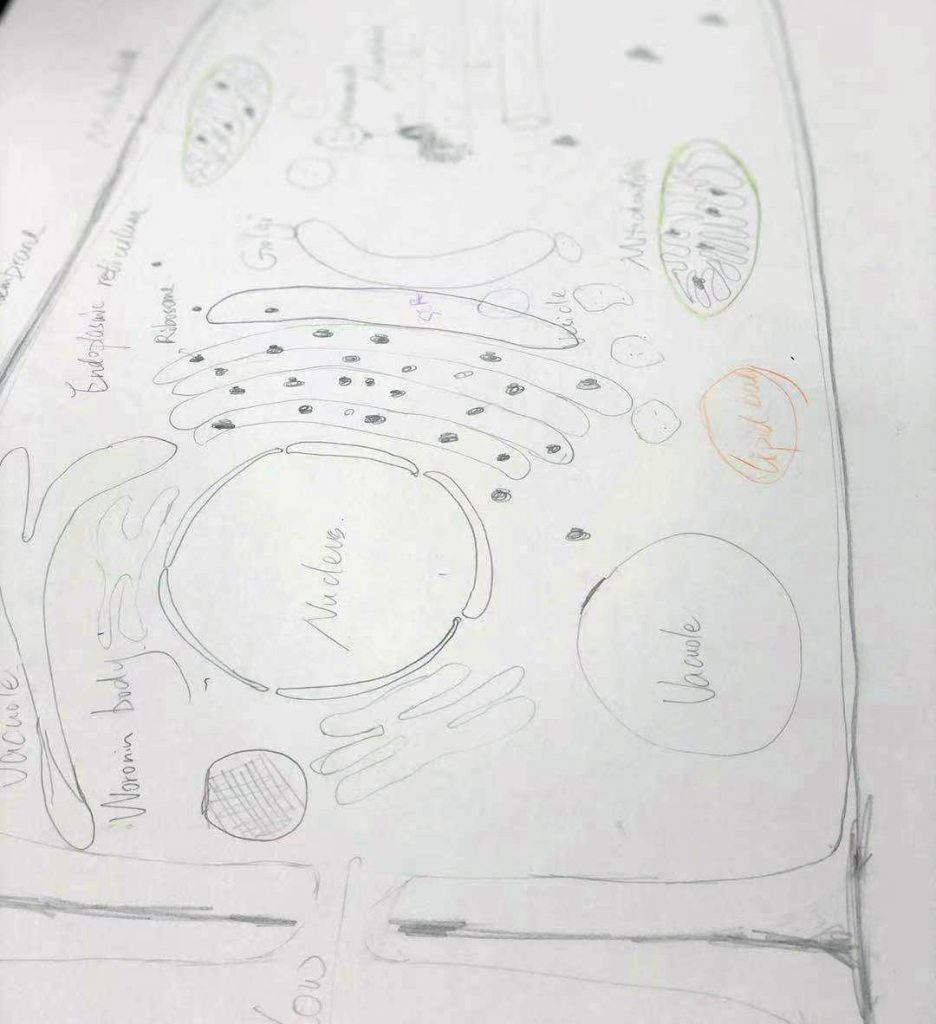

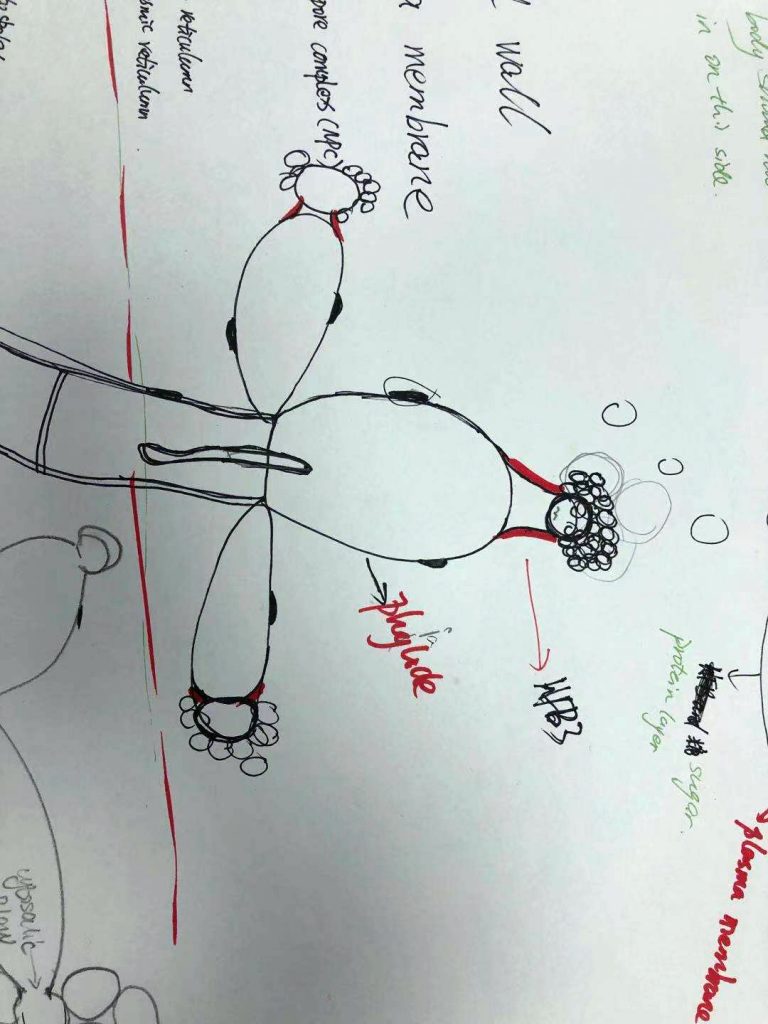

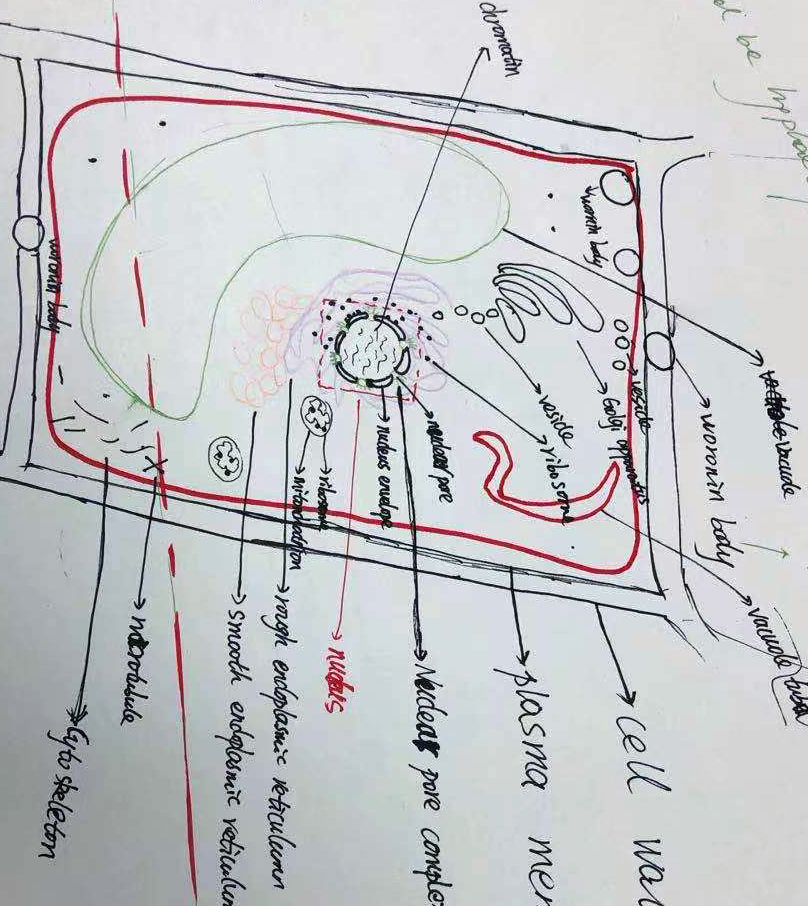

And then looked at the beautiful fungal cells:
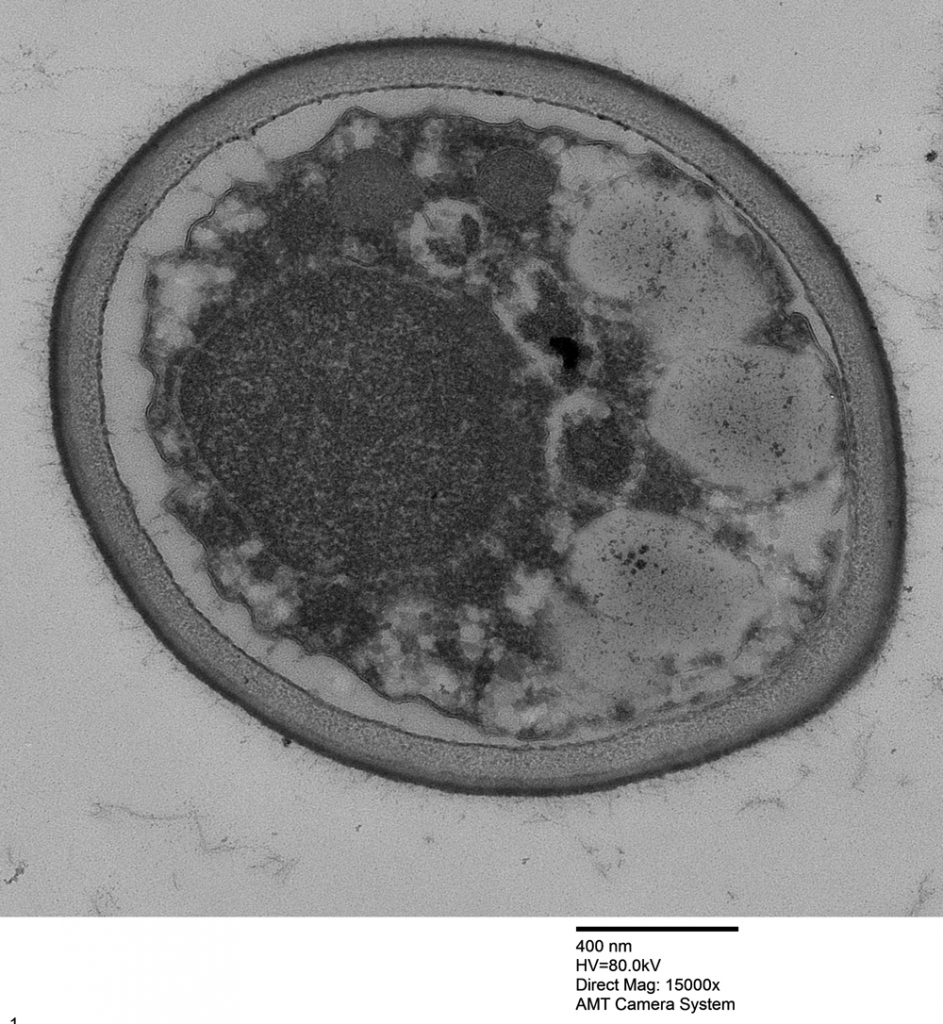
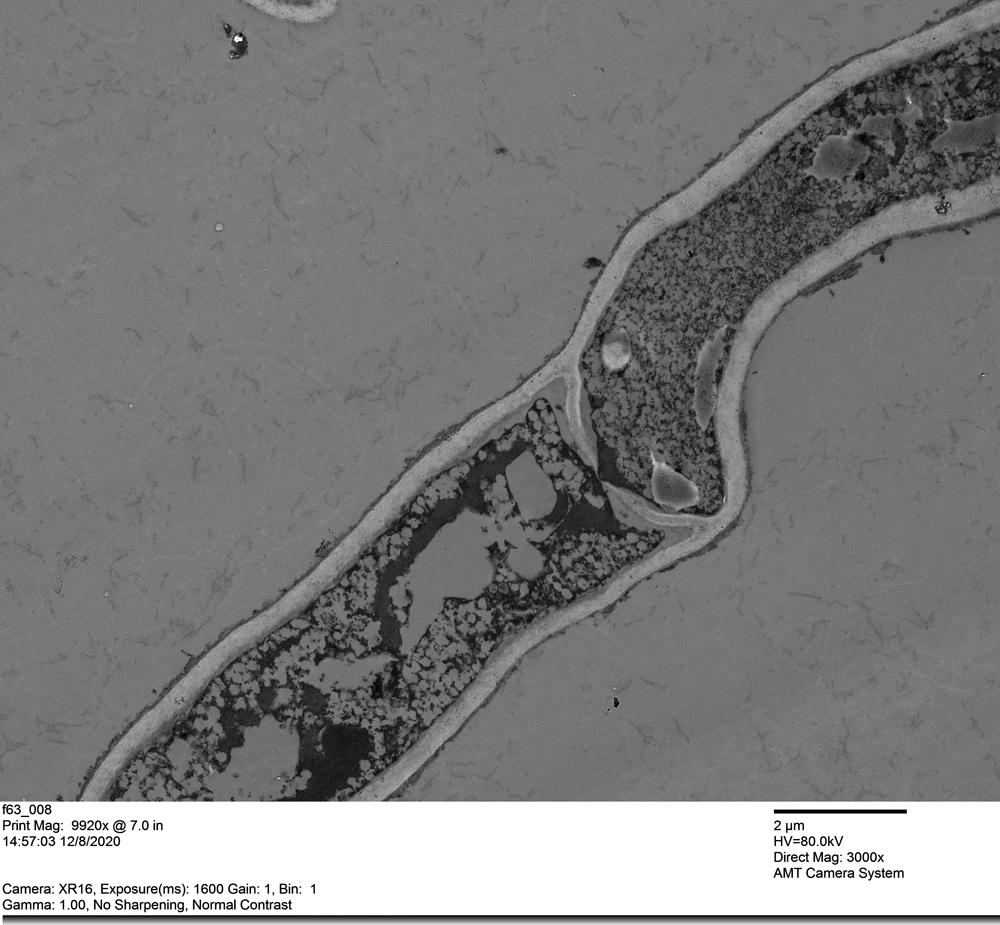
Dear Trichoderma Friends,
We wish you to be safe and have pleasant seasonal holidays. Let us hope that the new year will bring us luck that technology will win, we become stronger and more caring about each other and nature.
Merry Xmas and Happy New Year to you all!
