A complete list of publications may also be found at https://www.researchgate.net/profile/Irina_Druzhinina/contributions
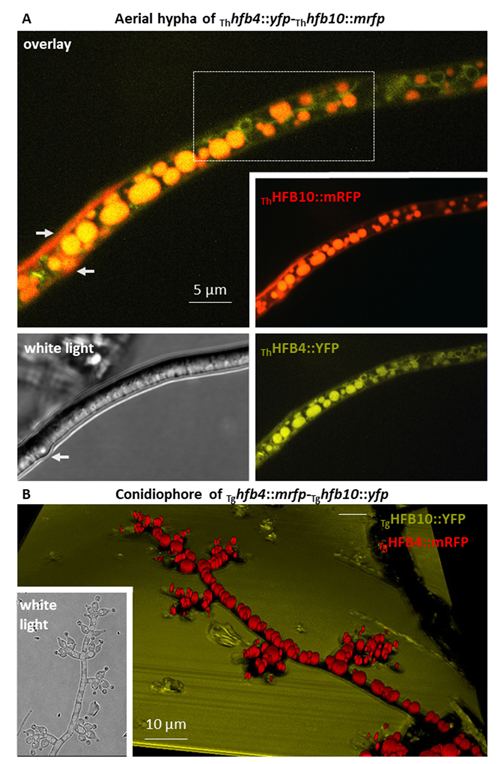
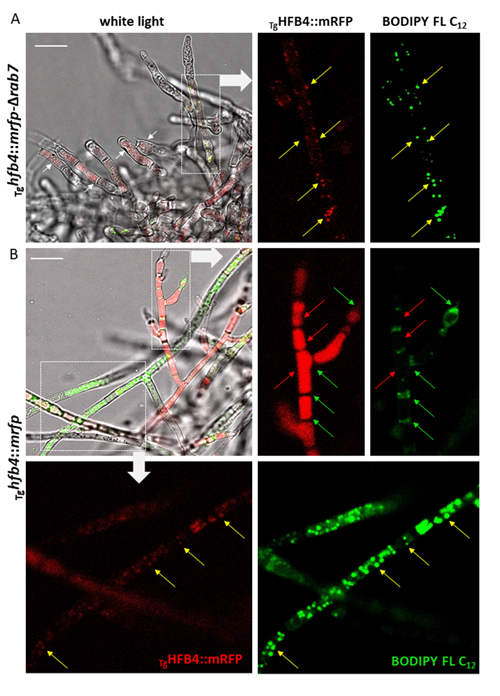
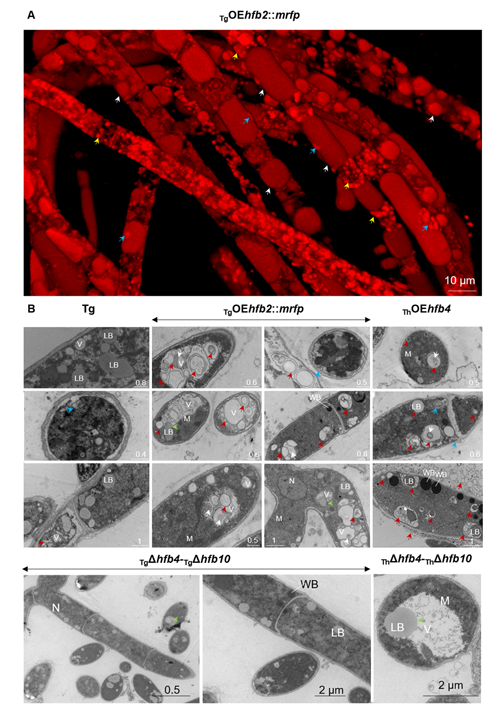
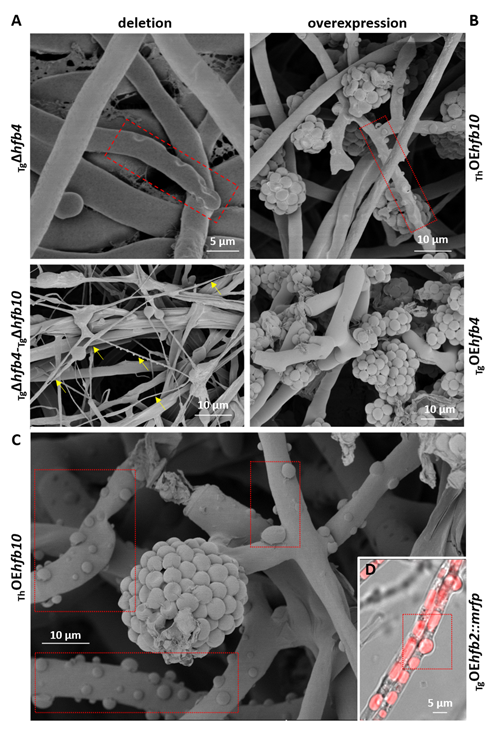
Cai, F., Zhao, Z., Gao R., Ding, M., Jiang, S., Gao, Q., Chenthamara, K., Grujic, M., Bayram-Akcapinar, G., Shen, Q., and Druzhinina, I. S. Intracellular hydrophobin vesicles modulate the architecture and longevity of fungal colonies. (preprint) doi: 10.1101/2020.08.18.255406
Cai F, Druzhinina IS (2020) In honor of John Bissett: Authoritative guidelines on molecular identification of Trichoderma. Fungal Diversity. (in press) doi: 10.1007/s13225-020-00464-4
Cai F, Gao R, Zhao Z, Ding M, Jiang S, Yagtu C, Zhu H, Zhang J, Ebner T, Mayrhofer-Reinhartshuber M, Kainz P, Chenthamara K, Akcapinar GB, Shen Q, Druzhinina IS (2020) Evolutionary compromises in fungal fitness: hydrophobins can hinder the adverse dispersal of conidiospores and challenge their survival. The ISME Journal 14 (10):2610-2624. doi:10.1038/s41396-020-0709-0
Ding MY, Chen W, Ma XC, Lv BW, Jiang SQ, Yu YN, Rahimi MJ, Gao RW, Zhao Z, Cai F, Druzhinina IS (2020) Emerging salt marshes as a source of Trichoderma arenarium sp. nov. and other fungal bio effectors for bio saline agriculture. Journal of Applied Microbiology :jam.14751. doi:10.1111/jam.14751
Gao R, Ding M, Jiang S, Zhao Z, Chenthamara K, Shen Q, Cai F, Druzhinina IS (2020) The Evolutionary and Functional Paradox of Cerato-platanins in Fungi. Appl Environ Microb 86 (13):e00696-00620, /aem/00686/00613/AEM.00696-00620.atom. doi:10.1128/AEM.00696-20
Hoenigsberger M, Pretzer C, Rahimi MJ, Kopchinskiy AG, Parich A, Laciny A, Metscher B, Chan CM, Lim LBL, Salim KA, Zettel H, Druzhinina IS, Schuhmacher R (2020) Strong antimicrobial and low insecticidal activity of mandibular gland reservoir content in Bornean “exploding ants” Colobopsis explodens Laciny & Zettel, 2018 (Hymenoptera: Formicidae). doi:10.25849/ MYRMECOL.NEWS_030:201
Lücking R, Aime MC, Robbertse B, Miller AN, Ariyawansa HA, Aoki T, Cardinali G, Crous PW, Druzhinina IS, Geiser DM, Hawksworth DL, Hyde KD, Irinyi L, Jeewon R, Johnston PR, Kirk PM, Malosso E, May TW, Meyer W, Öpik M, Robert V, Stadler M, Thines M, Vu D, Yurkov AM, Zhang N, Schoch CL (2020) Unambiguous identification of fungi: where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus 11 (1):14. doi:10.1186/s43008-020-00033-z
Pérez-Llano Y, Rodríguez-Pupo EC, Druzhinina IS, Chenthamara K, Cai F, Gunde-Cimerman N, Zalar P, Gostinčar C, Kostanjšek R, Folch-Mallol JL, Batista-García RA, Sánchez-Carbente MdR (2020) Stress Reshapes the Physiological Response of Halophile Fungi to Salinity. Cells 9 (3):525. doi:10.3390/cells9030525
Wijayawardene N, Hyde KD, Dai DQ, Tang LZ, Aptroot A, CastanedaRuiz RF, Druzhinina IS, Cai F, Ekanayaka AH, Erdogdu M (2020) A dynamic portal for a community-driven, continuously updated classification of Fungi and fungus-like organisms: outlineoffungi.org. Mycosphere 11 (1):1514-1526. doi:10.5943/mycosphere/11/1/11
Grujić M, Dojnov B, Potočnik I, Atanasova L, Duduk B, Srebotnik E, Druzhinina IS, Kubicek CP, Vujčić Z (2019) Superior cellulolytic activity of Trichoderma guizhouense on raw wheat straw. World Journal of Microbiology and Biotechnology 35 (12):194. doi:10.1007/s11274-019-2774-y
Hatvani L, Homa M, Chenthamara K, Cai F, Kocsubé S, Atanasova L, Mlinaric-Missoni E, Manikandan P, Revathi R, Dóczi I, Bogáts G, Narendran V, Büchner R, Vágvölgyi C, Druzhinina IS, Kredics L (2019) Agricultural systems as potential sources of emerging human mycoses caused by Trichoderma: a successful, common phylotype of Trichoderma longibrachiatum in the frontline. FEMS Microbiology Letters 366 (21):fnz246. doi:10.1093/femsle/fnz246
Hoenigsberger M, Kopchinskiy A, Bueschl C, Parich A, Laciny A, Zettel H, Salim K, Lim L, Druzhinina I, Schuhmacher R (2019) Volatiles from the Mandibular Gland Reservoir Content of Colobopsis explodens Laciny and Zettel, 2018, Worker Ants (Hymenoptera: Formicidae). Molecules 24 (19):3468. doi:10.3390/molecules24193468
Kubicek CP, Steindorff AS, Chenthamara K, Manganiello G, Henrissat B, Zhang J, Cai F, Kopchinskiy AG, Kubicek EM, Kuo A, Baroncelli R, Sarrocco S, Noronha EF, Vannacci G, Shen Q, Grigoriev IV, Druzhinina IS (2019) Evolution and comparative genomics of the most common Trichoderma species. BMC Genomics 20 (1):485. doi:10.1186/s12864-019-5680-7
Laciny A, Nemeschkal HL, Zettel H, Metscher B, Druzhinina IS (2019) Caste-specific morphological modularity in the ant tribe Camponotini (Hymenoptera, Formicidae). BMC Zoology 4 (1):9. doi:10.1186/s40850-019-0048-7
Marik T, Tyagi C, Balázs D, Urbán P, Szepesi Á, Bakacsy L, Endre G, Rakk D, Szekeres A, Andersson MA, Salonen H, Druzhinina IS, Vágvölgyi C, Kredics L (2019) Structural Diversity and Bioactivities of Peptaibol Compounds From the Longibrachiatum Clade of the Filamentous Fungal Genus Trichoderma. Front Microbiol 10:1434. doi:10.3389/fmicb.2019.01434
Zhang J, Miao Y, Rahimi MJ, Zhu H, Steindorff A, Schiessler S, Cai F, Pang G, Chenthamara K, Xu Y, Kubicek CP, Shen Q, Druzhinina IS (2019) Guttation capsules containing hydrogen peroxide: an evolutionarily conserved NADPH oxidase gains a role in wars between related fungi. Environmental Microbiology 21 (8):2644-2658. doi:10.1111/1462-2920.14575
Chadha S, Mehetre ST, Bansal R, Kuo A, Aerts A, Grigoriev IV, Druzhinina IS, Mukherjee PK (2018) Genome-wide analysis of cytochrome P450s of Trichoderma spp.: annotation and evolutionary relationships. Fungal Biology and Biotechnology 5 (1):12. doi:10.1186/s40694-018-0056-3
Druzhinina IS, Chenthamara K, Zhang J, Atanasova L, Yang D, Miao Y, Rahimi MJ, Grujic M, Cai F, Pourmehdi S, Salim KA, Pretzer C, Kopchinskiy AG, Henrissat B, Kuo A, Hundley H, Wang M, Aerts A, Salamov A, Lipzen A, LaButti K, Barry K, Grigoriev IV, Shen Q, Kubicek CP (2018) Massive lateral transfer of genes encoding plant cell wall-degrading enzymes to the mycoparasitic fungus Trichoderma from its plant-associated hosts. PLOS Genetics 14 (4):e1007322. doi:10.1371/journal.pgen.1007322
du Plessis IL, Druzhinina IS, Atanasova L, Yarden O, Jacobs K (2018) The diversity of Trichoderma species from soil in South Africa, with five new additions. Mycologia 110 (3):559-583. doi:10.1080/00275514.2018.1463059
Hoenigsberger M, Kopchinskiy AG, Parich A, Hiller K, Laciny A, Zettel H, Lim LBL, Salim KA, Druzhinina IS, Schuhmacher R (2018) Isolation of Mandibular Gland Reservoir Contents from Bornean ‘Exploding Ants’ (Formicidae) for Volatilome Analysis by GC-MS and Metabolite Detector. JoVE (138):57652. doi:10.3791/57652
Kamravamanesh D, Kovacs T, Pflügl S, Druzhinina I, Kroll P, Lackner M, Herwig C (2018) Increased poly-β-hydroxybutyrate production from carbon dioxide in randomly mutated cells of cyanobacterial strain Synechocystis sp. PCC 6714: Mutant generation and characterization. Bioresource Technology 266:34-44. doi:10.1016/j.biortech.2018.06.057
Laciny A, Zettel H, Kopchinskiy A, Pretzer C, Pal A, Abu Salim K, Javad Rahimi M, Hoenigsberger M, Lim L, Jaitrong W, Druzhinina IS (2018) Colobopsis explodens sp. n., model species for studies on “exploding ants” (Hymenoptera, Formicidae), with biological notes and first illustrations of males of the Colobopsis cylindrica group. Zookeys 751:1-40. doi:10.3897/zookeys.751.22661
Zettel H, Laciny A, Weeyawat J, Syaukani S, Kopchinskiy A, Druzhinina IS (2018) Evidence of predation in two species of the Colobopsis cylindrica group (Hymenoptera: Formicidae: Camponotini). Asian Myrmecology 10: e010011 (1-11) doi:10.20362/AM.010011
Druzhinina IS, Kubicek CP (2017) Genetic engineering of Trichoderma reesei cellulases and their production. Microb Biotechnol 10 (6):1485-1499. doi:10.1111/1751-7915.12726
Gaskell J, Kersten P, Larrondo LF, Canessa P, Martinez D, Hibbett D, Schmoll M, Kubicek CP, Martinez AT, Yadav J, Master E, Magnuson JK, Yaver D, Berka R, Lail K, Chen C, LaButti K, Nolan M, Lipzen A, Aerts A, Riley R, Barry K, Henrissat B, Blanchette R, Grigoriev IV, Cullen D (2017) Draft genome sequence of a monokaryotic model brown-rot fungus Postia (Rhodonia) placenta SB12. Genomics Data 14:21-23. doi:10.1016/j.gdata.2017.08.003
Laciny A, Zettel H, Metscher B, Kamariah AS, Kopchinskiy A, Druzhinina IS (2017) Morphological variation and mermithism in female castes of Colobopsis sp. nrSA, a Bornean “exploding ant” of the Colobopsis cylindrica group (Hymenoptera: Formicidae). Myrmecological News 24:91-106
Marik T, Urbán P, Tyagi C, Szekeres A, Leitgeb B, Vágvölgyi M, Manczinger L, Druzhinina IS, Vágvölgyi C, Kredics L (2017) Diversity Profile and Dynamics of Peptaibols Produced by Green Mould Trichoderma Species in Interactions with Their Hosts Agaricus bisporus and Pleurotus ostreatus. Chem Biodiversity 14 (6):e1700033. doi:10.1002/cbdv.201700033
Miao Y, Li P, Li G, Liu D, Druzhinina IS, Kubicek CP, Shen Q, Zhang R (2017) Two degradation strategies for overcoming the recalcitrance of natural lignocellulosic xylan by polysaccharides-binding GH10 and GH11 xylanases of filamentous fungi: Lignocellulosic xylan degradation strategies. Environmental Microbiology 19 (3):1054-1064. doi:10.1111/1462-2920.13614
Pretzer C, Druzhinina IS, Amaro C, Benediktsdóttir E, Hedenström I, Hervio‐Heath D, Huhulescu S, Schets FM, Farnleitner AH, Kirschner AKT (2017) High genetic diversity of Vibrio cholerae in the European lake Neusiedler See is associated with intensive recombination in the reed habitat and the long‐distance transfer of strains. Environmental Microbiology 19 (1):328-344. doi:10.1111/1462-2920.13612
Przylucka A, Akcapinar GB, Bonazza K, Mello-de-Sousa TM, Mach-Aigner AR, Lobanov V, Grothe H, Kubicek CP, Reimhult E, Druzhinina IS (2017) Comparative physiochemical analysis of hydrophobins produced in Escherichia coli and Pichia pastoris. Colloids and Surfaces B: Biointerfaces 159:913-923. doi:10.1016/j.colsurfb.2017.08.058
Przylucka A, Akcapinar GB, Chenthamara K, Cai F, Grujic M, Karpenko J, Livoi M, Shen Q, Kubicek CP, Druzhinina IS (2017) HFB7 – A novel orphan hydrophobin of the Harzianum and Virens clades of Trichoderma, is involved in response to biotic and abiotic stresses. Fungal Genetics and Biology 102:63-76. doi:10.1016/j.fgb.2017.01.002
Qin Y, Pan X, Kubicek C, Druzhinina I, Chenthamara K, Labbé J, Yuan Z (2017) Diverse Plant-Associated Pleosporalean Fungi from Saline Areas: Ecological Tolerance and Nitrogen-Status Dependent Effects on Plant Growth. Front Microbiol 8. doi:10.3389/fmicb.2017.00158
Davidson DW, Kopchinskiy A, Salim KA, Grujic M, Lim L, Mei CC, Jones TH, Casamatta D, Atanasova L, Druzhinina IS (2016) Nutrition of Borneo’s ‘exploding’ ants (Hymenoptera: Formicidae: Colobopsis ): a preliminary assessment. Biotropica 48 (4):518-527. doi:10.1111/btp.12323
de Man TJB, Stajich JE, Kubicek CP, Teiling C, Chenthamara K, Atanasova L, Druzhinina IS, Levenkova N, Birnbaum SSL, Barribeau SM, Bozick BA, Suen G, Currie CR, Gerardo NM (2016) Small genome of the fungus Escovopsis weberi, a specialized disease agent of ant agriculture. Proceedings of the National Academy of Sciences 113 (13):3567-3572. doi:10.1073/pnas.1518501113
Druzhinina IS, Kopchinskiy AG, Kubicek EM, Kubicek CP (2016) A complete annotation of the chromosomes of the cellulase producer Trichoderma reesei provides insights in gene clusters, their expression and reveals genes required for fitness. Biotechnology for Biofuels 9 (1):75. doi:10.1186/s13068-016-0488-z
Druzhinina IS, Kubicek CP (2016) Familiar Stranger. Ecological Genomics of the Model Saprotroph and Industrial Enzyme Producer Trichoderma reesei Breaks the Stereotypes In: Advances in Applied Microbiology, Adv Appl Microbiol. 2016;95:69-147.doi: 10.1016/bs.aambs.2016.02.001.
Druzhinina IS, Kubicek EM, Kubicek CP (2016) Several steps of lateral gene transfer followed by events of ‘birth-and-death’ evolution shaped a fungal sorbicillinoid biosynthetic gene cluster. BMC Evolutionary Biology 16 (1):269. doi:10.1186/s12862-016-0834-6
Laciny A, Zettel H, Druzhinina I (2016) Workers, soldiers, and gynes – morphometric characterization and description of the female castes of Camponotus singularis (Smith, 1858) (Hymenoptera, Formicidae). DEZ 63 (2):183-193. doi:10.3897/dez.63.9435
Qin Y, Druzhinina IS, Pan X, Yuan Z (2016) Microbially Mediated Plant Salt Tolerance and Microbiome-based Solutions for Saline Agriculture. Biotechnology Advances 34 (7):1245-1259. doi:10.1016/j.biotechadv.2016.08.005
Yuan Z, Druzhinina IS, Labbé J, Redman R, Qin Y, Rodriguez R, Zhang C, Tuskan GA, Lin F (2016) Specialized Microbiome of a Halophyte and its Role in Helping Non-Host Plants to Withstand Salinity. Scientific Reports: 2016 Aug 30;6:32467. doi: 10.1038/srep32467.
Zhang N, Yang D, Kendall JRA, Borriss R, Druzhinina IS, Kubicek CP, Shen Q, Zhang R (2016) Comparative Genomic Analysis of Bacillus amyloliquefaciens and Bacillus subtilis Reveals Evolutional Traits for Adaptation to Plant-Associated Habitats. Front Microbiol 7. doi:10.3389/fmicb.2016.02039
Bonazza K, Gaderer R, Neudl S, Przylucka A, Allmaier G, Druzhinina IS, Grothe H, Friedbacher G, Seidl-Seiboth V (2015) The fungal cerato-platanin protein EPL1 forms highly ordered layers at hydrophobic/hydrophilic interfaces. Soft Matter 11 (9):1723-1732. doi:10.1039/C4SM02389G
Ribitsch D, Herrero Acero E, Przylucka A, Zitzenbacher S, Marold A, Gamerith C, Tscheließnig R, Jungbauer A, Rennhofer H, Lichtenegger H, Amenitsch H, Bonazza K, Kubicek CP, Druzhinina IS, Guebitz GM (2015) Enhanced Cutinase-Catalyzed Hydrolysis of Polyethylene Terephthalate by Covalent Fusion to Hydrophobins. Appl Environ Microb 81 (11):3586-3592. doi:10.1128/AEM.04111-14
Yang D, Pomraning K, Kopchinskiy A, Karimi Aghcheh R, Atanasova L, Chenthamara K, Baker SE, Zhang R, Shen Q, Freitag M, Kubicek CP, Druzhinina IS (2015) Genome Sequence and Annotation of Trichoderma parareesei, the Ancestor of the Cellulase Producer Trichoderma reesei. Genome Announcements 3 (4):e00885-00815. doi:10.1128/genomeA.00885-15
Zhang J, Akcapinar GB, Atanasova L, Rahimi MJ, Przylucka A, Yang D, Kubicek CP, Zhang R, Shen Q, Druzhinina IS (2015) The neutral metallopeptidase NMP1 of Trichoderma guizhouense is required for mycotrophy and selfdefence. Environmental Microbiology: https://doi.org/10.1111/1462-2920.12966
Hori C, Ishida T, Igarashi K, Samejima M, Suzuki H, Master E, Ferreira P, Ruiz-Dueñas FJ, Held B, Canessa P, Larrondo LF, Schmoll M, Druzhinina IS, Kubicek CP, Gaskell JA, Kersten P, St. John F, Glasner J, Sabat G, Splinter BonDurant S, Syed K, Yadav J, Mgbeahuruike AC, Kovalchuk A, Asiegbu FO, Lackner G, Hoffmeister D, Rencoret J, Gutiérrez A, Sun H, Lindquist E, Barry K, Riley R, Grigoriev IV, Henrissat B, Kües U, Berka RM, Martínez AT, Covert SF, Blanchette RA, Cullen D (2014) Analysis of the Phlebiopsis gigantea Genome, Transcriptome and Secretome Provides Insight into Its Pioneer Colonization Strategies of Wood. PLoS Genetics 10 (12):e1004759. doi:10.1371/journal.pgen.1004759
Karimi Aghcheh R, Németh Z, Atanasova L, Fekete E, Paholcsek M, Sándor E, Aquino B, Druzhinina IS, Karaffa L, Kubicek CP (2014) The VELVET A Orthologue VEL1 of Trichoderma reesei Regulates Fungal Development and Is Essential for Cellulase Gene Expression. PLoS ONE 9 (11):e112799. doi:10.1371/journal.pone.0112799
Pfliegler WP, Atanasova L, Karanyicz E, Sipiczki M, Bond U, Druzhinina IS, Sterflinger K, Lopandic K (2014) Generation of New Genotypic and Phenotypic Features in Artificial and Natural Yeast Hybrids.12
Agneheh RK, Druzhinina IS, Kubicek CP (2013) The Putative Protein Methyltransferase LAE1 of Trichoderma atroviride Is a Key Regulator of Asexual Development and Mycoparasitism. PLoS ONE 8 (6):e67144. doi:10.1371/journal.pone.0067144
Atanasova L, Crom SL, Gruber S, Coulpier F, Seidl-Seiboth V, Kubicek CP, Druzhinina IS (2013) Comparative transcriptomics reveals different strategies of Trichoderma mycoparasitism. BMC Genomics 14 (1):121. doi:10.1186/1471-2164-14-121
Atanasova L, Knox BP, Kubicek CP, Druzhinina IS, Baker SE (2013) The Polyketide Synthase Gene pks4 of Trichoderma reesei Provides Pigmentation and Stress Resistance. Eukaryotic Cell 12 (11):1499-1508. doi:10.1128/EC.00103-13
Erper I, Turkkan M, Atanasova L, Druzhinina IS, Karaca GH, CebI-KilIcoglu M (2013) Integrated assessment of the mycoparasitic and phytostimulating properties of Trichoderma strains against Rhizoctonia solani. Bulgarian Journal of Agricultural Science 19(4):737-743
Espino-Rammer L, Ribitsch D, Przylucka A, Marold A, Greimel KJ, Herrero Acero E, Guebitz GM, Kubicek CP, Druzhinina IS (2013) Two Novel Class II Hydrophobins from Trichoderma spp. Stimulate Enzymatic Hydrolysis of Poly(Ethylene Terephthalate) when Expressed as Fusion Proteins. Appl Environ Microb 79 (14):4230-4238. doi:10.1128/AEM.01132-13
Karaffa L, Coulier L, Fekete E, Overkamp KM, Druzhinina IS, Mikus M, Seiboth B, Novák L, Punt PJ, Kubicek CP (2013) The intracellular galactoglycome in Trichoderma reesei during growth on lactose. Applied Microbiology and Biotechnology 97 (12):5447-5456. doi:10.1007/s00253-012-4667-y
Lehner SM, Atanasova L, Neumann NKN, Krska R, Lemmens M, Druzhinina IS, Schuhmacher R (2013) Isotope-Assisted Screening for Iron-Containing Metabolites Reveals a High Degree of Diversity among Known and Unknown Siderophores Produced by Trichoderma spp. Appl Environ Microb 79 (1):18-31. doi:10.1128/AEM.02339-12
López-Quintero CA, Atanasova L, Franco-Molano AE, Gams W, Komon-Zelazowska M, Theelen B, Müller WH, Boekhout T, Druzhinina I (2013) DNA barcoding survey of Trichoderma diversity in soil and litter of the Colombian lowland Amazonian rainforest reveals Trichoderma strigosellum sp. nov. and other species. Antonie van Leeuwenhoek 104 (5):657-674. doi:10.1007/s10482-013-9975-4
Mulaw T, Druzhinina I, Kubicek C, Atanasova L (2013) Novel Endophytic Trichoderma spp. Isolated from Healthy Coffea arabica Roots are Capable of Controlling Coffee Tracheomycosis. Diversity 5 (4):750-766. doi:10.3390/d5040750
Pummer BG, Atanasova L, Bauer H, Bernardi J, Druzhinina IS, Fröhlich-Nowoisky J, Grothe H (2013) Spores of many common airborne fungi reveal no ice nucleation activity in oil immersion freezing experiments. Biogeosciences 10 (12):8083-8091. doi:10.5194/bg-10-8083-2013
Degenkolb T, Karimi Aghcheh R, Dieckmann R, Neuhof T, Baker SE, Druzhinina IS, Kubicek CP, Brückner H, von Döhren H (2012) The Production of Multiple Small Peptaibol Families by Single 14-Module Peptide Synthetases in Trichoderma/Hypocrea. Chem Biodiversity 9 (3):499-535. doi:10.1002/cbdv.201100212
Druzhinina IS, Komoń-Zelazowska M, Ismaiel A, Jaklitsch W, Mullaw T, Samuels GJ, Kubicek CP (2012) Molecular phylogeny and species delimitation in the section Longibrachiatum of Trichoderma. Fungal Genetics and Biology 49 (5):358-368. doi:10.1016/j.fgb.2012.02.004
Druzhinina IS, Shelest E, Kubicek CP (2012) Novel traits of Trichoderma predicted through the analysis of its secretome. FEMS Microbiology Letters 337 (1):1-9. doi:10.1111/j.1574-6968.2012.02665.x
Friedl MA, Druzhinina IS (2012) Taxon-specific metagenomics of Trichoderma reveals a narrow community of opportunistic species that regulate each other’s development. Microbiology 158 (1):69-83. doi:10.1099/mic.0.052555-0
Mukherjee PK, Buensanteai N, Moran-Diez ME, Druzhinina IS, Kenerley CM (2012) Functional analysis of non-ribosomal peptide synthetases (NRPSs) in Trichoderma virens reveals a polyketide synthase (PKS)/NRPS hybrid enzyme involved in the induced systemic resistance response in maize. Microbiology 158 (1):155-165. doi:10.1099/mic.0.052159-0
Samuels GJ, Ismaiel A, Mulaw TB, Szakacs G, Druzhinina IS, Kubicek CP, Jaklitsch WM (2012) The Longibrachiatum Clade of Trichoderma: a revision with new species. Fungal Diversity 55 (1):77-108. doi:10.1007/s13225-012-0152-2
Druzhinina IS, Seidl-Seiboth V, Herrera-Estrella A, Horwitz BA, Kenerley CM, Monte E, Mukherjee PK, Zeilinger S, Grigoriev IV, Kubicek CP (2011) Trichoderma: the genomics of opportunistic success. Nature Reviews Microbiology 9 (10):749-759. doi:10.1038/nrmicro2637
Gal-Hemed I, Atanasova L, Komon-Zelazowska M, Druzhinina IS, Viterbo A, Yarden O (2011) Marine Isolates of Trichoderma spp. as Potential Halotolerant Agents of Biological Control for Arid-Zone Agriculture. Appl Environ Microb 77 (15):5100-5109. doi:10.1128/AEM.00541-11
Kubicek CP, Herrera-Estrella A, Seidl-Seiboth V, Martinez DA, Druzhinina IS, Thon M, Zeilinger S, Casas-Flores S, Horwitz BA, Mukherjee PK, Mukherjee M, Kredics L, Alcaraz LD, Aerts A, Antal Z, Atanasova L, Cervantes-Badillo MG, Challacombe J, Chertkov O, McCluskey K, Coulpier F, Deshpande N, von Döhren H, Ebbole DJ, Esquivel-Naranjo EU, Fekete E, Flipphi M, Glaser F, Gómez-Rodríguez EY, Gruber S, Han C, Henrissat B, Hermosa R, Hernández-Oñate M, Karaffa L, Kosti I, Le Crom S, Lindquist E, Lucas S, Lübeck M, Lübeck PS, Margeot A, Metz B, Misra M, Nevalainen H, Omann M, Packer N, Perrone G, Uresti-Rivera EE, Salamov A, Schmoll M, Seiboth B, Shapiro H, Sukno S, Tamayo-Ramos JA, Tisch D, Wiest A, Wilkinson HH, Zhang M, Coutinho PM, Kenerley CM, Monte E, Baker SE, Grigoriev IV (2011) Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biology 12 (4):R40. doi:10.1186/gb-2011-12-4-r40
Atanasova L, Druzhinina IS (2010) Global nutrient profiling by Phenotype MicroArrays: a tool complementing genomic and proteomic studies in conidial fungi. J Zhejiang Univ Sci B 11 (3):151-168. doi:10.1631/jzus.B1000007
Atanasova L, Jaklitsch WM, Komoń-Zelazowska M, Kubicek CP, Druzhinina IS (2010) Clonal Species Trichoderma parareesei sp. nov. Likely Resembles the Ancestor of the Cellulase Producer Hypocrea jecorina/T. reesei. Appl Environ Microb 76 (21):7259-7267. doi:10.1128/AEM.01184-10
Druzhinina IS, Komoń-Zelazowska M, Atanasova L, Seidl V, Kubicek CP (2010) Evolution and Ecophysiology of the Industrial Producer Hypocrea jecorina (Anamorph Trichoderma reesei) and a New Sympatric Agamospecies Related to It. PLoS ONE 5 (2):e9191. doi:10.1371/journal.pone.0009191
Druzhinina IS, Kubicek CP, Komon-Zelazowska M, Belayneh Mulaw T, Bissett J (2010) The Trichoderma harzianum demon: complex speciation history resulting in coexistence of hypothetical biological species, recent agamospecies and numerous relict lineages. BMC Evolutionary Biology 10 (1):94. doi:10.1186/1471-2148-10-94
Mulaw TB, C.P. K, Druzhinina IS (2010) The Rhizosphere of Coffea Arabica in Its Native Highland Forests of Ethiopia Provides a Niche for a Distinguished Diversity of Trichoderma. Diversity 2(4)
Paz Z, Komon-Zelazowska M, Druzhinina IS, Aveskamp MM, Shnaiderman A, Aluma Y, Carmeli S, Ilan M, Yarden O (2010) Diversity and potential antifungal properties of fungi associated with a Mediterranean sponge. Fungal Diversity 42 (1):17-26. doi:10.1007/s13225-010-0020-x
Kemptner J, Marchetti-Deschmann M, Mach R, Druzhinina IS, Kubicek CP, Allmaier G (2009) Evaluation of matrix-assisted laser desorption/ionization (MALDI) preparation techniques for surface characterization of intact Fusarium spores by MALDI linear time-of-flight mass spectrometry. Rapid Commun Mass Spectrom 23 (6):877-884. doi:10.1002/rcm.3949
Kredics Ls, Kocsubé Sn, Nagy Ls, Komoń-Zelazowska M, Manczinger Ls, Sajben E, Nagy A, Vágvölgyi C, Kubicek CP, Druzhinina IS, Hatvani Lrn (2009) Molecular identification of Trichoderma species associated with Pleurotus ostreatus and natural substrates of the oyster mushroom. FEMS Microbiology Letters 300 (1):58-67. doi:10.1111/j.1574-6968.2009.01765.x
Le Crom S, Schackwitz W, Pennacchio L, Magnuson JK, Culley DE, Collett JR, Martin J, Druzhinina IS, Mathis H, Monot F, Seiboth B, Cherry B, Rey M, Berka R, Kubicek CP, Baker SE, Margeot A (2009) Tracking the roots of cellulase hyperproduction by the fungus Trichoderma reesei using massively parallel DNA sequencing. Proceedings of the National Academy of Sciences 106 (38):16151-16156. doi:10.1073/pnas.0905848106
Migheli Q, Balmas V, Komoñ-Zelazowska M, Scherm B, Fiori S, Kopchinskiy AG, Kubicek CP, Druzhinina IS (2009) Soils of a Mediterranean hot spot of biodiversity and endemism (Sardinia, Tyrrhenian Islands) are inhabited by pan-European, invasive species of Hypocrea/Trichoderma. Environmental Microbiology 11 (1):35-46. doi:10.1111/j.1462-2920.2008.01736.x
Mikus M, Hatvani Lrn, Neuhof T, Komoń-Zelazowska M, Dieckmann R, Schwecke T, Druzhinina IS, von D�hren H, Kubicek CP (2009) Differential Regulation and Posttranslational Processing of the Class II Hydrophobin Genes from the Biocontrol Fungus Hypocrea atroviridis. Appl Environ Microb 75 (10):3222-3229. doi:10.1128/AEM.01764-08
Zachow C, Berg C, Müller H, Meincke R, Komon-Zelazowska M, Druzhinina IS, Kubicek CP, Berg G (2009) Fungal diversity in the rhizosphere of endemic plant species of Tenerife (Canary Islands): relationship to vegetation zones and environmental factors. The ISME Journal 3 (1):79-92. doi:10.1038/ismej.2008.87
Brunner K, Omann M, Pucher ME, Delic M, Lehner SM, Domnanich P, Kratochwill K, Druzhinina I, Denk D, Zeilinger S (2008) Trichoderma G protein-coupled receptors: functional characterisation of a cAMP receptor-like protein from Trichoderma atroviride. Current Genetics 54 (6):283-299. doi:10.1007/s00294-008-0217-7
Druzhinina IS, Komoń-Zelazowska M, Kredics L, Hatvani L, Antal Z, Belayneh T, Kubicek CP (2008) Alternative reproductive strategies of Hypocrea orientalis and genetically close but clonal Trichoderma longibrachiatum, both capable of causing invasive mycoses of humans. Microbiology 154 (11):3447-3459. doi:10.1099/mic.0.2008/021196-0
Friedl MA, Kubicek CP, Druzhinina IS (2008) Carbon Source Dependence and Photostimulation of Conidiation in Hypocrea atroviridis. Appl Environ Microb 74 (1):245-250. doi:10.1128/AEM.02068-07
Friedl MA, Schmoll M, Kubicek CP, Druzhinina IS (2008) Photostimulation of Hypocrea atroviridis growth occurs due to a cross-talk of carbon metabolism, blue light receptors and response to oxidative stress. Microbiology 154(Pt 4):1229-41
Jaklitsch WM, Kubicek CP, Druzhinina IS (2008) Three European species of Hypocrea with reddish brown stromata and green ascospores. Mycologia 100 (5):796-815. doi:10.3852/08-039
Kubicek CP, Baker SE, Gamauf C, Kenerley CM, Druzhinina IS (2008) Purifying selection and birth-and-death evolution in the class II hydrophobin gene families of the ascomycete Trichoderma/Hypocrea. BMC Evolutionary Biology 8 (1):4. doi:10.1186/1471-2148-8-4
Kubicek CP, Komon-Zelazowska M, Druzhinina IS (2008) Fungal genus Hypocrea/Trichoderma: from barcodes to biodiversity. J Zhejiang Univ Sci B 9 (10):753-763. doi:10.1631/jzus.B0860015
Seidl V, Gamauf C, Druzhinina IS, Seiboth B, Hartl L, Kubicek CP (2008) The Hypocrea jecorina (Trichoderma reesei) hypercellulolytic mutant RUT C30 lacks a 85 kb (29 gene-encoding) region of the wild-type genome. BMC Genomics 9 (1):327. doi:10.1186/1471-2164-9-327
Váczy KZ, Sándor E, Karaffa L, Fekete E, Fekete É, Árnyasi M, Czeglédi L, Kövics GJ, Druzhinina IS, Kubicek CP (2008) Sexual Recombination in the Botrytis cinerea Populations in Hungarian Vineyards. Phytopathology 98 (12):1312-1319. doi:10.1094/PHYTO-98-12-1312
Druzhinina IS, LaFe K, Willinger B, Komoń-Zelazowska M, Ammirati J, Kubicek CP, Rogers JD (2007) An Unknown Hypocreaceae Species Isolated from Human Lung Tissue of a Patient with Non-Fatal Pulmonary Fibrosis. Clinical Microbiology Newsletter 29 (23):180-184. doi:10.1016/j.clinmicnews.2007.11.002
Hatvani L, Antal Z, Manczinger L, Szekeres A, Druzhinina IS, Kubicek CP, Nagy A, Nagy E, Vágvölgyi C, Kredics L (2007) Green Mold Diseases of Agaricus and Pleurotus spp. Are Caused by Related but Phylogenetically Different Trichoderma Species. Phytopathology 97 (4):532-537. doi:10.1094/PHYTO-97-4-0532
Komoń-Zelazowska M, Bissett J, Zafari D, Hatvani L, Manczinger L, Woo S, Lorito M, Kredics L, Kubicek CP, Druzhinina IS (2007) Genetically Closely Related but Phenotypically Divergent Trichoderma Species Cause Green Mold Disease in Oyster Mushroom Farms Worldwide. Appl Environ Microb 73 (22):7415-7426. doi:10.1128/AEM.01059-07
Komon-Zelazowska M, Neuhof T, Dieckmann R, von Döhren H, Herrera-Estrella A, Kubicek CP, Druzhinina IS (2007) Formation of Atroviridin by Hypocrea atroviridis Is Conidiation Associated and Positively Regulated by Blue Light and the G Protein GNA3. Eukaryotic Cell 6 (12):2332-2342. doi:10.1128/EC.00143-07
Kubicek CP, Komoń-Zelazowska M, Sándor E, Druzhinina IS (2007) Facts and Challenges in the Understanding of the Biosynthesis of Peptaibols by Trichoderma. Chem Biodiversity 4 (6):1068-1082. doi:10.1002/cbdv.200790097
Nagy V, Seidl V, Szakacs G, Komoń-Zelazowska M, Kubicek CP, Druzhinina IS (2007) Application of DNA Bar Codes for Screening of Industrially Important Fungi: the Haplotype of Trichoderma harzianum Sensu Stricto Indicates Superior Chitinase Formation. Appl Environ Microb 73 (21):7048-7058. doi:10.1128/AEM.00995-07
Neuhof T, Dieckmann R, Druzhinina IS, Kubicek CP, Nakari-Setälä T, Penttilä M, von Döhren H (2007) Direct identification of hydrophobins and their processing in Trichoderma using intact-cell MALDI-TOF MS: Trichoderma hydrophobins. FEBS Journal 274 (3):841-852. doi:10.1111/j.1742-4658.2007.05636.x
Neuhof T, Dieckmann R, Druzhinina IS, Kubicek CP, von Döhren H (2007) Intact-cell MALDI-TOF mass spectrometry analysis of peptaibol formation by the genus Trichoderma/Hypocrea: can molecular phylogeny of species predict peptaibol structures? Microbiology 153 (10):3417-3437. doi:10.1099/mic.0.2007/006692-0
Schuster A, Kubicek CP, Friedl MA, Druzhinina IS, Schmoll M (2007) Impact of light on Hypocrea jecorina and the multiple cellular roles of ENVOY in this process. BMC Genomics 8 (1):449. doi:10.1186/1471-2164-8-449
Zhang C-l, Liu S-p, Lin F-c, Kubicek CP, Druzhinina IS (2007) Trichoderma taxi sp. nov., an endophytic fungus from Chinese yew Taxus mairei. FEMS Microbiology Letters 270 (1):90-96. doi:10.1111/j.1574-6968.2007.00659.x
Dela Cruz TEE, Schulz BE, Kubicek CP, Druzhinina IS (2006) Carbon source utilization by the marine Dendryphiella species D. arenaria and D. salina. FEMS Microbiology Ecology 58 (3):343-353. doi:10.1111/j.1574-6941.2006.00184.x
Druzhinina IS, Kopchinskiy AG, Kubicek CP (2006) The first 100 Trichoderma species characterized by molecular data. Mycoscience 47 (2):55-64. doi:10.1007/S10267-006-0279-7
Druzhinina IS, Schmoll M, Seiboth B, Kubicek CP (2006) Global Carbon Utilization Profiles of Wild-Type, Mutant, and Transformant Strains of Hypocrea jecorina. Appl Environ Microb 72 (3):2126-2133. doi:10.1128/AEM.72.3.2126-2133.2006
Jaklitsch WM, Komon M, Kubicek CP, Druzhinina IS (2006) Hypocrea crystalligena sp. nov., a common European species with a white-spored Trichoderma anamorph. Mycologia 98(3):499-513
Jaklitsch WM, Komon M, Kubicek CP, Druzhinina IS (2006) Hypocrea voglmayrii sp. nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia 97(6):1365-78
Jaklitsch WM, Samuels GJ, Dodd SL, Lu B-S, Druzhinina IS (2006) Hypocrea rufa/Trichoderma viride: a reassessment, and description of five closely related species with and without warted conidia. Studies in Mycology 56:135-177. doi:10.3114/sim.2006.56.04
Samuels GJ, Dodd SL, Lu B-S, Petrini O, Schroers H-J, Druzhinina IS (2006) The Trichoderma koningii aggregate species. Studies in Mycology 56:67-133. doi:10.3114/sim.2006.56.03
Seidl V, Druzhinina IS, Kubicek CP (2006) A screening system for carbon sources enhancing β-N-acetylglucosaminidase formation in Hypocrea atroviridis (Trichoderma atroviride). Microbiology 152 (7):2003-2012. doi:10.1099/mic.0.28897-0
Druzhinina I, Kubicek CP (2005) Species concepts and biodiversity in Trichoderma and Hypocrea: from aggregate species to species clusters? J Zhejiang Univ Sci B (2):100-112. doi:10.1631/jzus.2005.B0100
Druzhinina IS, Kopchinskiy AG, Komoń M, Bissett J, Szakacs G, Kubicek CP (2005) An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. Fungal Genetics and Biology 42 (10):813-828. doi:10.1016/j.fgb.2005.06.007
Kopchinskiy A, Komoń M, Kubicek CP, Druzhinina IS (2005) TrichoBlast: A Multilocus Database for Trichoderma and Hypocrea Identifications. Mycol Res 109 (6):658-660. doi:10.1017/S0953756205233397
Zhang C-l, Druzhinina IS, Kubicek CP, Xu T (2005) Trichoderma biodiversity in China: Evidence for a North to South distribution of species in East Asia. FEMS Microbiology Letters 251 (2):251-257. doi:10.1016/j.femsle.2005.08.034
Druzhinina I, Palma-Oliveira JM (2004) Radioactive contamination of wild mushrooms: a cross-cultural risk perception study. Journal of Environmental Radioactivity 74 (1-3):83-90. doi:10.1016/j.jenvrad.2004.01.025
Druzhinina IS, Chaverri P, Fallah P, Kubicek CP, Samuels GJ (2004) Hypocrea flaviconidia, a new species from Costa Rica with yellow conidia. Studies in Mycology, 50:401–407.
Gherbawy Y, Druzhinina I, Shaban GM, Wuczkowsky M, Yaser M, El-Naghy MA, Prillinger H-J, Kubicek CP (2004) Trichoderma populations from alkaline agricultural soil in the Nile valley, Egypt, consist of only two species. Mycol Prog 3 (3):211-218. doi:10.1007/s11557-006-0091-y
Kraus GF, Druzhinina I, Gams W, Bissett J, Zafari D, Szakacs G, Koptchinski A, Prillinger H, Zare R, Kubicek CP (2004) Trichoderma brevicompactum sp. nov. Mycologia 96 (5):1059-1073. doi:10.1080/15572536.2005.11832905
Lu B, Druzhinina IS, Fallah P, Chaverri P, Gradinger C, Kubicek CP, Samuels GJ (2004) Hypocrea/Trichoderma species with pachybasium-like conidiophores: teleomorphs for T. minutisporum and T. polysporum and their newly discovered relatives. Mycologia 96(2):310-42
Pail M, Peterbauer T, Seiboth B, Hametner C, Druzhinina I, Kubicek CP (2004) The metabolic role and evolution of l-arabinitol 4-dehydrogenase of Hypocrea jecorina. Eur J Biochem 271 (10):1864-1872. doi:10.1111/j.1432-1033.2004.04088.x
Bissett J, Szakacs G, Nolan CA, Druzhinina I, Gradinger C, Kubicek CP (2003) New species of Trichoderma from Asia. Can J Bot 81 (6):570-586. doi:10.1139/b03-051
Kubicek CP, Bissett J, Druzhinina I, Kullnig-Gradinger C, Szakacs G (2003) Genetic and metabolic diversity of Trichoderma: a case study on South-East Asian isolates. Fungal Genetics and Biology 38 (3):310-319. doi:10.1016/S1087-1845(02)00583-2
Wuczkowski M, Druzhinina I, Gherbawy Y, Klug B, Prillinger H, Kubicek CP (2003) Species pattern and genetic diversity of Trichoderma in a mid-European, primeval floodplain-forest. Microbiological Research 158 (2):125-133. doi:10.1078/0944-5013-00193
Druzhinina IS, Insarova ID, Shnyreva AV, D’Yakov YT, Altukhov YP (1997) Homokaryotic mycelial pellets of basidiomycetes: Application to genetic analysis. Russian Journal of Genetics 33 (5):521-526